This is an old revision of the document!
Table of Contents
Frequently asked questions: General
Error messages
Negative standard errors
Standard errors in WOMBAT are derived a) assuming large samples, and b) a series of approximations - see the Technical Details section of the manual. In some cases, this approximation simply fails. Reasons for this may be, for instance, that your sample is very small, or that you are dealing with a model which is overparameterised, which includes multivariate analyses where some covariance matrices have eigenvalues which are effectively zero. In the latter scenario, it may help to fit a reduced rank model. Otherwise, you simply have to accept that the approximation of standard errors does not always work.
Crash "Segmentation fault"
- Q: WOMBAT crashes with a “Segmentation fault” with a heap of unintelligible numbers to follow – this seems to happen quite early on, i.e. during the set-up phase before any likelihoods are printed to the screen. What is wrong?
- A: Long answer on separate page – here
Message "Small/Invalid Pivot"
- Q: When I run WOMBAT I sometimes get a message to the screen, saying either small, negative or invalid pivot - What does it mean and what should I do about it?
- A: Long answer on separate page – here
Message "lnkloc" : dimension exceeded !!
- Q: WOMBAT stops with this error message – why does this occur and, more importantly, is there anything I can do to make this go away (other than reducing the size of my analysis)?
- A: Depending on what exactly you are trying to do, this message can appear in different parts of the program – it is basically a 'safety catch' making sure your analysis has enough room. Most arrays needed are allocated at exactly the size required for the analysis; however, in some spots WOMBAT guesses at the space needed and, occasionally, this guess is simply wrong. In addition, there are some hard-coded maximum values and if your model of analysis is very big it may exceed one of these values.
- The first place this message may occur is during calculation of the inverse of the numerator relationship matrix (I have never seen this happen though); if it does, you are likely to have a very big and very dense pedigree and would be best advised to reduce the size of your analysis.
- The second possibility is that you are trying to use the PX-EM algorithm (this is the default for the first few iterates when there are more than 18 parameters to be estimated, so you may be doing this without having explicitly chosen to do so), and WOMBAT stops before the first iterate. WOMBAT has a guess on how many non-zero off-diagonal elements there are in one triangle of the coefficient matrix of the mixed model equations and then attempts to set this up and to determine the actual number. If this is too small, it will stop. A particular scenario when this has been found to occur it the case of an analysis trying to estimate a covariance matrix of reduced rank (option PC), where the rank is substantially less than the dimension of the matrix.
To get around this, you could try to:- use the AI-REML algorithm from the start (see run time option
—-aireml), especially if you have good starting values - set the initial guess manually using the run time option
—-choozhz
- The third place this can happen is when using the iterative solutions module; again, setting the maximum number manually via the
—-choozhzoption may help.
Confusion
Zero residual covariances
- Q: My multivariate analysis gives `estimates' of residual covariances which are zero, and
SumModel.outtells me that WOMBAT thinks that there are no animals which have records for the corresponding pairs of traits – but I know that there are animals which do. Why is WOMBAT so confused ? - A: WOMBAT requires the data file to be in a certain order to correctly identify that an individual has multiple records – please check the manual! Perhaps you have forgotten to sort the data file, or the sort did not work properly. Please sort the data file as required and try again – and double-check that
SumModel.outgives the correct numbers of individuals with records for each pair of traits before proceeding to the estimation step.
Another possibility is that you have specified starting values of zero for the residual covariances – this causes WOMBAT not to estimate these values and fix them at their starting values. However, in that caseSumModel.outshould report numbers of individuals with pairs of records greater than zero.
Interaction for fixed effects
- Q: I want to fit a model with an interaction between two fixed effects. How do I do that ? The manual says Not Yet implemented – surely, that can't be true ?
- A: The manual is slightly misleading here: Yes, of course you can fit a model with an interaction. What the manual means is that WOMBAT won't to the relevant coding for you. So what you have to do is:
- Generate a code for the interaction from the two fixed effects codes and write this to a new column in your data file. For example, if fixed effect A has a maximum value of 8571 and you want to fit an interaction between fixed effects A and B, the new code could be 10000*code(B)+code(A). In doing so, ensure that your new code is still a valid integer variable, i.e. does not exceed 2,147,483,647.
- Fit this new effect as if it were a cross-classified fixed effect.
- Check carefully for any dependencies among the fixed effects before running your analysis. If necessary, tell WOMBAT about them using the
ZEROUToption. This is particularly important, if you want to fit effects A (or B) in addition to the interaction - you then have the interaction nested in A (or B), i.e. if effect A has a levels, you have (a-1) additional dependencies among the fixed effects. WOMBAT may find some of them, but you cannot rely on it. ~~UP~~
- Q: I am fitting age as a quadratic covariable in the model, using the default polynomial regression. WOMBAT gives me estimates of regression coefficients in
FixSolutions.out, but: a) I can't find the intercept, and b) these estimates don't make sense, i.e. when I plug them into the regression equation, I don't get a curve which fits the data. What is wrong? - A: Long answer on separate page – here ~~UP~~
- Q: What options does WOMBAT provide for the analysis of categorical traits – I cannot find anything about it in the manual.
- A: None – the manual clearly states that WOMBAT is geared towards the analysis of continuous traits (only!), assuming a multivariate normal distribution. ~~UP~~
What is ?
Pruning and pedigree reduction
- Q: What exactly does WOMBAT do to the pedigree information I supply?
- A: WOMBAT tries to `condense' the pedigrees supplied as far as possible without loosing any information – this makes computations more efficient. There are two separate processes:
- Pedigree reduction: In this step WOMBAT simply scans the data and pedigree files and eliminates any records from the pedigree file for individuals which are not in any way connected to an individual in the data. These do not supply any information – if they were left in the analysis, their estimated breeding values would be zero. The pedigree file for the remaining individuals is written out the file ''ReducedPed.dat''. There is no run time option to switch this step off.
- Pruning: There may be other individuals which do not contribute information (for variance component estimation) – these are individuals without records and a pedigree link to only one other individual. However, through the pedigree link, there is some information on their genetic value, i.e. if left in the analysis, their breeding value estimates would be non-zero. If one of these occurs as a sire or dam, WOMBAT replaces them with an 'unknown' code and eliminates them from the list of pedigree records. This `pruning' is done recursively, until all such individuals are eliminated, and is done both from the top of the pedigree down (oldest to youngest) and from the bottom up (youngest to oldest). Pruning can be switched off using the
−−noprunerun time option. ~~UP~~
How-To
- Q: I want to fit a covariable, but there are some records where this is not recorded. What should I do? WOMBAT doesn't seem to allow for a 'missing value' code there, so I have set them all to a value of zero. Is that o.k. ?
- A: No, it is not! Strictly speaking, you should delete all records for which you don't have the covariable recorded. WOMBAT will not do that for you! Setting 'missing values' to zero will create havouc (unless the mean covariable in the data is zero), as WOMBAT will treat those as observed values and therefore may estimate regression coefficient which don't fit the data at all. In some instances, you may not be willing to delete these records - if you really must retain them, you should set the value for the missing covariable to the mean of the covariable in the data. This would yield an analysis where you adjust some records for the covariable, but not others; hence you should not do this for covariables which have a substantial effect on the variance of your observations. ~~UP~~
- Q: I am dealing with a trait subject to maternal effects. However, the identity of the dam is not recorded for a lot of animals, and WOMBAT won't let me run an analysis fitting maternal effects if the data contain dam codes of zero. What can I do to analyse this trait properly?
- A: Sadly, WOMBAT cannot generate information out of nothing, i.e. in order to estimate an effect you need to record it. You could:
- Fit a sire instead of an animal model. This should at least give you an estimate of the additive genetic variance unbiased by maternal effects.
- Fit an animal model for the subset of the data which do have known dams, i.e. delete all records on animals with unidentified dams.
- If only a small proportion of dams are unknown (less than 5% or so), you could replace the missing dam codes by some dummy code, assigning a different fictitious dam to each animal with unknown dam.
N.B. Even if all animals in the data have known dams, you may have animals in the pedigree with unknown dams. If you wish to fit a maternal genetic effect, you will then similarly need to assign dummy dam codes for all unknown dams, i.e. the third column in the pedigree file cannot contain any zeros. Again, your analysis will work best if this proportion of dams is relatively small.
- Q: I have run a standard variance component analysis and found the solutions for effects fitted in the model. However, corresponding standard errors are missing - how do I get them?
- A: In a mixed model analysis, approximate lower bound standard errors are obtained from the diagonal elements of the inverse of the coefficient matrix. The default method for variance component estimation is the “average informatiom” algorithm. The implementation in WOMBAT does not involve inversion of the coefficient matrix - hence standard errors are not simply a by-product. You can enforce calculation of standard errors by simply adding the option
FORCE-SEin aSPECIALblock at the end of the parameter file.
SPECIAL FORCE-SE END
- Q: I am using WOMBAT to obtain breeding values for animals. How can I get the corresponding accuracies?
- A: Long answer on separate page – here ~~UP~~
- Q: I am trying to use WOMBAT to analyse data on plants where I can have selfing or individuals which can be both sires and dams. However, WOMBAT stops with an error message claiming that my pedigree records are invalid. What can I do?
- A: WOMBAT has been written with the analysis of data from livestock in mind where such pedigrees are indeed not plausible and such checks thus help to track pedigree errors. You can switch these checks off using a run time option of
−−self(not in the manual), provided that you are confident the standard rules to set up the inverse of the numerator relationship matrix are applicable. An alternative is to set up the inverse relationship matrix yourself and supply it as a generalized inverse covariance matrix (.ginfile).~~UP~~
- Q: I am fitting a random regression model with age as the `control' variable and want to calculate breeding value estimates for specific age points. How do I do that?
- A: WOMBAT will give you estimates of the random regression (RR) coefficients. Say you have fitted a random effect called
animalto represent direct, additive genetic effects – the estimates of the RR coefficients are the in the file ''RnSoln_animal.dat''. Further, say you have fitted 3 basis functions , such as the first three Legendre polynomials, and that you are interested in breeding values at age a. You then need to evaluate the regression equation
, such as the first three Legendre polynomials, and that you are interested in breeding values at age a. You then need to evaluate the regression equation 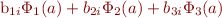 for each animal, with
for each animal, with 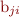 the estimated RR for individual i and
the estimated RR for individual i and 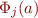 the j-th basis function evaluated for a. WOMBAT does not perform these simple calculations for you, but to make this task easier, it writes out a file with the basis functions evaluated for all values of the control variable occurring in the data – see the manual for details! ~~UP~~
the j-th basis function evaluated for a. WOMBAT does not perform these simple calculations for you, but to make this task easier, it writes out a file with the basis functions evaluated for all values of the control variable occurring in the data – see the manual for details! ~~UP~~
- Q: I want to run WOMBAT in `batch mode' - however it stops with a “Programmed pause” (and a warning message) and expects me to hit `Enter' to continue. How do I get around that?
- A: Easy, use run option
−−batchbut, please, read the warning messages!
- Q: WOMBAT writes out a number of binary (
.bin) files. Is there any way I can access the information in these - I'd be interested in things like the inbreeding coefficients for individual animals. - A: Well, these files are not really meant to be read by the outside world - that's one reason why they are binary. However, I have a small number of simple, stand-alone programs to read these. You need a FORTRAN compiler and a modicum of programming knowledge to modify them to generate output according to your requirements. See: Auxiliary Programs
NB: The current version of WOMBAT writes out the inbreeding coefficients (in %) for individual levels of genetic effects as the last column in the corresponding "RnSoln_*.dat" files.~~UP~~
- Q: I am trying to run a multivariate analysis, but WOMBAT stops immediately with a message that that the
Matrix of starting values is “invalid”– what does this mean and what can I do about it? And why can't WOMBAT fix this automatically? - A: Long answer on separate page – here
~~UP~~
- Q: How do I run a weighted analysis in WOMBAT?
- A: That depends on what exactly you mean by `weighted'.
- Fixed weights: If your weights do not depend on the variance components to be estimated or any of the effects in the model of analysis, then really all you need to do is weigh your observations appropriately (e.g. multiply or divide by weight) prior to the analysis.
- Variable weights: In other cases, the weights to apply depend on the variance components and change with each round of iteration.
The best strategy to deal with this is to carry out your analysis one iterate at a time, scaling observations as required before each iterate – it may be a bit tedious, but it will do what you want and give you full flexibility in applying different kinds of weights. - WOMBAT does have an experimental SPECIAL option for a WEIGHTED analysis, but this is rather limited in what it does and has not been thoroughly tested; See WOMBAT manual. ~~UP~~
- Q: I want to fit a model with heterogeneous residual variances. Can WOMBAT do that? I can't seem to find anything in the manual.
- A: There is no direct option for heterogeneous variances for standard uni- and multivariate analyses. However, this does not mean you cannot fit such models. The trick is to think of your problem as a special kind of random regression analysis – if you can formulate it as such, it is highly likely that you'll be able to fit it in WOMBAT. ~~UP~~
- Q: I want to estimate a genetic covariance matrix with a factor-analytic structure, but I can't find the option to do so. I thought WOMBAT could do this kind of analysis.
- A: Yes, WOMBAT can do this. However, there is no explicit option for it. You cannot fit a standard, multivariate model and tell WOMBAT to impose a FA structure. Instead, you need to fit what is referred to as `extended' factor-analytic model – fitting a vector of common factors and a vector of specific effects as two separate random effects. See this page for further details.
~~UP~~
- Q: I am trying to run analyses where I estimate the genetic covariance matrix at reduced rank. I thought a good strategy might be to start with a full rank analysis and to reduce the rank one by one. Doing so, I thought I could use the estimates from the previous run as starting values. However, specifying a run time option of
-cto tell WOMBAT to pick up the starting values from the fileBestPointdoesn't work – the program ignores it completely and uses the values from the.parfile. Why? - A: This is indeed a very good strategy for such analyses. There is just one small step missing:
- If you look at the file
BestPointyou'll see two numbers on the first line, the log likelihood value and the number of parameters. - When a continuation run is specified, WOMBAT will read the latter number and compare it to the number determined from the model & type of analysis specified in the
.parfile. If the two don't match, the input fromBestPointis unceremoniously ignored. This is a small `safety catch' which helps avoiding input from an inappropriate file. - What that means is that you need to edit
BestPointand replace the old number of parameters with the number for the next reduced rank run. This is easy to work out: Say you have a genetic covariance matrix for q traits. The full rank estimate has p=q(q+1)/2 parameters, reducing the rank r by one reduces p by one, reducing r to q-2 reduces p by 3, reducing r to q-3 reduces p by 6, and so forth. The general formula for the number of parameters to be estimated in a matrix of size q and rank r is p = r(2q-r+1)/2 .~~UP~~
- Q: I would like to calculate p-values for estimates of fixed effects - but cannot find the appropriate degrees of freedom in the WOMBAT output. Where are they?
- A: Unfortunately, WOMBAT does not attempt to determine residual degrees of freedom for such tests.
~~UP~~
- Q: I want to run WOMBAT from within R - is that possible and how do I do it?
- A: Yes. Provided you have set up the relevant paths (so that R can find the WOMBAT executables) you can run WOMBAT like any other system command, e.g.
system2(“wombat”,args=c(“-v”,“wombat.par”)). Of course all the usual input files must exist and you need to make sure that R is looking for them in the correct place.
Strange Behaviour
Almost linear convergence
- Q: I am running an analysis using the average information algorithm. I thought this was supposed to converge quadratically once it got sufficiently close to the maximum of the likelihood. However, my analysis is taking many, many iterates, each yielding only a small improvement – an almost linear convergence pattern as one might expect from the EM-algorithm. Any idea what is going on or how to improve on this?
- A: There are a number of typical reasons for such behaviour
- You could have a `numerical' problem. For instance, you might have a trait with very large or very small variance. If so, some scaling might be helpful (see manual).
- You could have a data set which does not support estimation of the model you are trying to fit, i.e. a small data set or many parameters to be estimated. If you have a multivariate analysis with many traits, you might want to try a reduced rank analysis. For a random regression model, you might be able to reduce the order of fit or, again, reduce the rank of the covariance matrices to be estimated.
- Your analysis might suffer from an ill-conditioned average information (AI) matrix, i.e. an AI matrix with an eigenvalue close to zero (which often indicates an over-parameterized model!) or a matrix where the largest eigenvalue is very much larger than the smallest. If so, WOMBAT attempts some damage control by modifying the AI matrix. The default is to add a small constant to the diagonal elements. Sometimes the guess for this constant is too large, slowing down convergence. This occurs especially if there is one very large eigenvalue, e.g. in a multi-trait analysis with many traits, whether you are estimating covariance matrices at reduced rank or not.
Things to look at and do:- Check the norm of the gradient vector and the Newton decrement (columns 5 and 7 in the
Iteratesfile). Are they reasonably small, i.e. is your analysis indeed at a point where you would normally expect quadratic convergence? - Use the
-voption to get WOMBAT to print out the values of the extreme eigenvalues. Is the smallest eigenvalue safely non-zero but less than unity and the largest eigenvalue more than ten thousand times larger? If so, WOMBAT may treat the AI matrix as ill-conditioned. - Check column 8 in the
Iteratesfile. This gives the step size multiplier. I would expect this to be consistently 1.000. - Check the second last column in the
Iteratesfile – this gives the constant which is added automatically. - If this constant is non-zero, you could try making it smaller. For instance, the command line option
−−modaim2,0.5halves it,−−modaim2,0.1reduces it to one tenth of the default value. NB: If you decrease this too much, you are likely to get strange jumps in parameter estimates and log likelihood values or the algorithm might fail altogether. - Alternatively, try one of the other mechanisms available to deal with such an AI matrix which is not well-conditioned (see manual).
Miscellaneous
Mac version
- Q I am looking for a Mac version - why isn't there one?
- A To compile a Mac version, I would need access to a Mac computer - which I don't have. I am told that either the Windows or Linux versions run o.k. in something like a Virtualbox guest environment. I am cross-compiling the Windows version under Linux, but have not found the equivalent for Mac - if you come across a simple way to compile under Linux for a Mac target, let me know.
UPDATE: A kind visitor compiled WOMBAT on his MAC - this is available for download now. NB: This is a “one-off” and will not be updated, nor are bug fixes possible.
Who uses it?
- Q: Is anybody actually using WOMBAT? I have not seen any publications citing it - so it can't be any good.
- A: A number of people use it. Bear in mind that is was only reased in mid-2006. A small, but growing list of publications is here.
This list has last been updated sometime in early 2012.